Data Analysis
2D-Gel Spot Picking
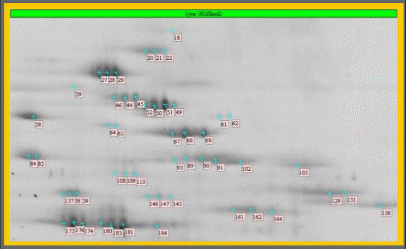
Detail of a Sypro Ruby stained large gel - sample: Synechocystis sp. 6803 thylakoids.
The sample, a subcellular membrane, exhibited a total of more than 1000 different protein species. About 100 functionally different gene expression products are known to be present in thylakoid membranes.The spots annotated with comment flags had been selected for spot picking.
(Courtesy of Dr. Irrgang, Max-Volmer-Institut, Technische Universität Berlin, Germany)
Depending on the project design chosen - differential display study vs. proteome inventory study, spot picking can for itself become a logistic issue. While differential display studies typically barely select more than 100 spots for protein identification, a proteome inventory study aiming to identify all spots present in a 20x30cm large gel, readily targets 5000 or more spots for picking.
Depending on the spot picker type as well as the actual physical parameters of a gel, a spot picker can do an 95%-99% errorfree job. However, some spots will simply remain in the gel, some will be lost on the way the spot picker tool travels between the gel and its spot collecting device. Although gels immobilised onto glass plates are routinely used for automated spot picking, local failure in gel adhesion can result in the loss of larger gel areas and the concomitant loss of so far unpicked spots. Minor local failures in gel adhesion may not necessarily be obvious before the start of the spot picking, however, even they can still contribute to a decrease of the success rate of picking.
As a consequence an approach aiming to give each of 5000 or more spots of a gel a good change during mass spectrometric protein identification, might require to actually pick 20000 or more spots (corresponding to 3-4 immobilized gels) in order to make sure that every spot has been picked twice. This, however, is often required to provide sufficient material for the succeeding protein identification.